| Class | Gene | Protein name | Sig. tests | Genbank ID | UniProt ID | Amino acid composition table | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| AA0 | POPTR_0001s04840 (EnsemblPlants link) | Peroxidase (EC 1.11.1.7) | 16 | AHL39107.1(NCBI query) | B9GLK7(UniProtKB link) | Amino Acid Composition(ExPASy link) Protein sequence(FASTA) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
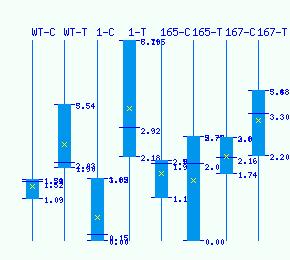
Normalized RNA-seq library counts for POPTR_0001s04840
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Gene | Protein name | Sig. tests | Genbank ID | UniProt ID | Amino acid composition table | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| AA0 | POPTR_0018s02910 (EnsemblPlants link) | Peroxidase (EC 1.11.1.7) | 4 | AHL39193.1(NCBI query) | B9IL94(UniProtKB link) | Amino Acid Composition(ExPASy link) Protein sequence(FASTA) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
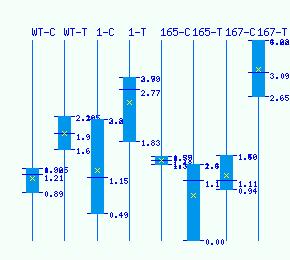
Normalized RNA-seq library counts for POPTR_0018s02910
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||